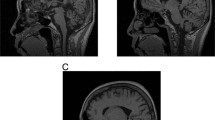
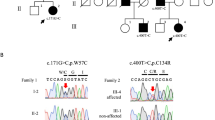
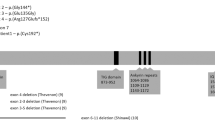
Abstract
Autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS or SACS) is an early onset neurodegenerative disease with high prevalence (carrier frequency 1/22) in the Charlevoix-Saguenay-Lac-Saint-Jean (CSLSJ) region of Quebec. We previously mapped the gene responsible for ARSACS to chromosome 13q11 and identified two ancestral haplotypes. Here we report the cloning of this gene, SACS, which encodes the protein sacsin. The ORF of SACS is 11,487 bp and is encoded by a single gigantic exon spanning 12,794 bp. This exon is the largest to be identified in any vertebrate organism. The ORF is conserved in human and mouse. The putative protein contains three large segments with sequence similarity to each other and to the predicted protein of an Arabidopsis thaliana ORF. The presence of heat-shock domains suggests a function for sacsin in chaperone-mediated protein folding. SACS is expressed in a variety of tissues, including the central nervous system. We identified two SACSmutations in ARSACS families that lead to protein truncation, consistent with haplotype analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bouchard, J.-P. Recessive spastic ataxia of Charlevoix-Saguenay. in Handbook of Clinical Neurology 16: Hereditary Neuropathies and Spinocerebellar Degenerations (ed. de Jong, J.M.B.V.) 451–459 (Elsevier, Amsterdam, 1991).
Bouchard, J.P. et al>. Autosomal recessive spastic ataxia of Charlevoix-Saguenay. Neuromuscul. Disord. 8, 474– 479 (1998).
Richter, A. et al. Location score and haplotype analyses of the locus for autosomal recessive spastic ataxia of Charlevoix-Saguenay in chromosome region 13q11. Am. J. Hum. Genet. 64, 768– 775 (1999).
Charbonneau, H. & Robert, N. The French origins of the Canadian population 1608–1759. in Historical Atlas of Canada Volume I: From the Beginning to 1800 (ed. Harris, R.C.) plate 45 (University of Toronto Press, Toronto, 1987).
Kruglyak, L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nature Genet. 22, 139– 144 (1999).
Hästbacka, J. et al. Linkage disequilibrium mapping in isolated founder populations: diastrophic dysplasia in Finland. Nature Genet. 2, 204–211 (1992).
Thompson, E.A. & Neel, J.V. Allelic disequilibrium and allele frequency distribution as a function of social and demographic history. Am. J. Hum. Genet. 60, 197–204 (1997).
Graham, J. & Thompson, E.A. Disequilibrium likelihoods for fine-scale mapping of a rare allele. Am. J. Hum. Genet. 63, 1517–1530 (1998).
Engert, J.C. et al. High resolution physical and transcript map of the autosomal recessive spastic ataxia of Charlevoix-Saguenay (ARSACS) candidate region in chromosome 13q11. Genomics 62, 156– 164 (1999).
Fink, A.L. Chaperone-mediated protein folding. Physiol. Rev. 79 , 425–449 (1999).
Buchner, J. Hsp90 & Co.—a holding for folding. Trends Biochem. Sci. 24, 136–141 (1999).
Gupta, R.S. Phylogenetic analysis of the 90 kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol. Biol. Evol. 12, 1063– 1073 (1995).
Nakai, K. & Kanehisa, M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14, 897–911 (1992).
McCarthy, L.C. et al. A first-generation whole-genome radiation hybrid map spanning the mouse genome. Genome Res. 7, 1153– 1161 (1997).
Van Etten, W.J. et al. Radiation hybrid map of the mouse genome. Nature Genet. 22, 384–387 ( 1999).
Brown, C.J. et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 71, 527–542 (1992).
Porchet, N., Aubert, J.P. & Laine, A. MUC5B, the 10.7-kb large central exon encodes various alternate subdomains resulting in a super-repeat. J. Biol. Chem. 272, 3168–3178 ( 1997).
Marcotte, E.M. et al. Detecting protein function and protein-protein interactions from genome sequences. Science 285, 751– 753 (1999).
Prodromou, C. et al. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90, 65–75 (1997).
Kimura, Y., Yahara, I. & Lindquist, S. Role of the protein chaperone YDJ1 in establishing Hsp90-mediated signal transduction pathways. Science 268, 1362–1365 (1995).
Dittmar, K.D., Banach, M., Galigniana, M.D. & Pratt, W.B. The role of DnaJ-like proteins in glucocorticoid receptor·hsp90 heterocomplex assembly by the reconstituted hsp90·p60·hsp70 foldosome complex. J. Biol. Chem. 273, 7358– 7366 (1998).
Cummings, C.J. et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nature Genet. 19, 148–154 ( 1998).
Dickie, M.M. Tumbler, tb. Mouse News Lett. 32 , 45 (1965).
De Braekeleer, M. Geographic distribution of 18 autosomal recessive disorders in the French Canadian population of Saguenay-Lac-St-Jean, Quebec. Ann. Hum. Biol. 22, 111–122 ( 1995).
Knoppers, B.M. & Laberge, C. DNA sampling and informed consent. CMAJ 144, 128– 129 (1991).
Birren, B.W., Mancino, V. & Shizuya, H. Bacterial artificial chromosomes. in Genome Analysis: A Laboratory Manual (eds Birren, B. et al .) 241– 295 (Cold Spring Harbor Laboratory Press, Plainview, 1999).
Bonfield, J.K., Smith, K.F. & Staden, R. A new DNA sequence assembly program. Nucleic Acids Res. 23, 4992–4999 (1995).
Bonfield, J.K. & Staden, R. Experiment files and their application during large-scale sequencing projects. DNA Seq. 6, 109–117 ( 1996).
Boysen, C., Simon, M.I. & Hood, L. Fluorescence-based sequencing directly from bacterial and P1-derived artificial chromosomes. Biotechniques 23, 978–982 (1997).
Smith, R.F., Wiese, B.A., Wojzynski, M.K., Davison, D.B. & Worley, K.C. BCM search launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res. 6, 454 –462 (1996).
Schalling, M. et al. Neuropeptide Y and catecholamine synthesizing enzymes and their mRNAs in rat sympathetic neurons and adrenal glands: studies on expression, synthesis and axonal transport after pharmacological and experimental manipulations using hybridization techiques and radioimmunoassay. Neuroscience 41, 753–766 ( 1991).
Weeks, D.E., Sobel, E., O'Connell, J.R. & Lange, K. Computer programs for multilocus haplotyping of general pedigrees. Am. J. Hum. Genet. 56, 1506–1507 (1995).
Austerlitz, F. & Heyer, E. Impact of demographic distribution and population growth rate on haplotypic diversity linked to a disease gene and their consequences for the estimation of recombination rate: example of a French Canadian population. Genet. Epidemiol. 16, 2–14 (1999 ).
Devlin, B. & Risch, N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics 29, 311–322 (1995).
de la Chapelle, A. & Wright, F.A.D. Linkage disequilibrium mapping in isolated populations: the example of Finland revisited. Proc. Natl Acad. Sci. USA 95, 12416– 12423 (1998).
Boehnke, M. Limits of resolution of genetic linkage studies: implications for the positional cloning of human disease genes. Am. J. Hum. Genet. 55, 379–390 (1994).
Graham, J. Disequilibrium fine-mapping a rare allele via coalesent models of gene ancestry. Thesis, Univ. Washington (1998).
Austerlitz, F. & Heyer, E. Social transmission of reproductive behavior increases frequency of inherited disorders in a young-expanding population. Proc. Natl Acad. Sci. USA 95, 15140–15144 (1998).
Heyer, E. One founder/one gene hypothesis in a new expanding population: Saguenay (Quebec, Canada). Hum. Biol. 71, 99– 109 (1999).
McNally, E.M. et al. Mild and severe muscular dystrophy caused by a single γ-sarcoglycan mutation. Am. J. Hum. Genet. 59, 1040– 1047 (1996).
Nagase, T. et al. Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 5, 277 –286 (1998).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680 (1994).
Acknowledgements
We thank the patients and families for participation; J. Graham, E.A. Thompson, B.F.F. Ouellette, J.D. Rioux, T.M. Fujiwara, P. Lee, M. Shapiro and C.M. Neville for helpful discussions; E.A. Thompson for a computer program for the coalescent method; E. Heyer and H. Vézina for demographic information; J. Ma for assistance in the creation of M13 subclone libraries; F. Gosselin and C. Prevost for sample collection and pedigree information; C. Goguen for administrative assistance; and C. Bieri for computer expertise. This work was supported by grants from the Medical Research Council of Canada (MRC; to A.R., S.B.M., K.M. and T.J.H.), the Muscular Dystrophy Association of Canada (to A.R., S.B.M. and T.J.H.), the March of Dimes (to A.R. and S.B.M.), the National Ataxia Foundation (to A.R. and S.B.M.), l'Institut Interuniversitaire de Recherches sur les Populations (to J.M., T.J.H. and K.M.), the Canadian Genetic Diseases Network (Networks of Centres of Excellence Program (NCE); to T.J.H. and K.M.), the Mathematics of Information Technology and Complex Systems (NCE; to K.M.), the Swedish Medical Research Council (to M.S.) and a research contract from Bristol-Myers Squibb, Millennium Pharmaceuticals Inc. and Affymetrix (to E.S.L. and T.J.H.). T.J.H. is a recipient of a Clinician Scientist Award from the MRC.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Engert, J., Bérubé, P., Mercier, J. et al. ARSACS, a spastic ataxia common in northeastern Québec, is caused by mutations in a new gene encoding an 11.5-kb ORF. Nat Genet 24, 120–125 (2000). https://doi.org/10.1038/72769
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/72769
This article is cited by
-
Widening the clinical, radiological and genetic spectrum of autosomal recessive ataxia of Charlevoix-Saguenay in Indian patients
Acta Neurologica Belgica (2024)
-
A SACS deletion variant in Great Pyrenees dogs causes autosomal recessive neuronal degeneration
Human Genetics (2023)
-
Natural History of Autosomal Recessive Spastic Ataxia of Charlevoix-Saguenay: a 4-Year Longitudinal Study
The Cerebellum (2023)
-
Transcriptome analysis of gene expression profiling from the deep sea in situ to the laboratory for the cold seep mussel Gigantidas haimaensis
BMC Genomics (2022)
-
Documenting manifestations and impacts of autosomal recessive spastic ataxia of Charlevoix–Saguenay to develop patient-reported outcome
Orphanet Journal of Rare Diseases (2022)